A multi-scale approach to understanding the mechanisms of mobile DNA driven antimicrobial resistance transmission
Interventions
Surveillance
Transmission
- Orsola Barabas, European Molecular Biology Laboratory, Germany (Coordinator)
- Peer Bork, European Molecular Biology Laboratory, Germany (Partner)
- Maria Fällman, Umeå University, Sweden (Partner)
- Johan Normark, Norrlands University Hospital, Sweden (Partner)
- Gerard Wright, McMaster University, Canada (Partner)
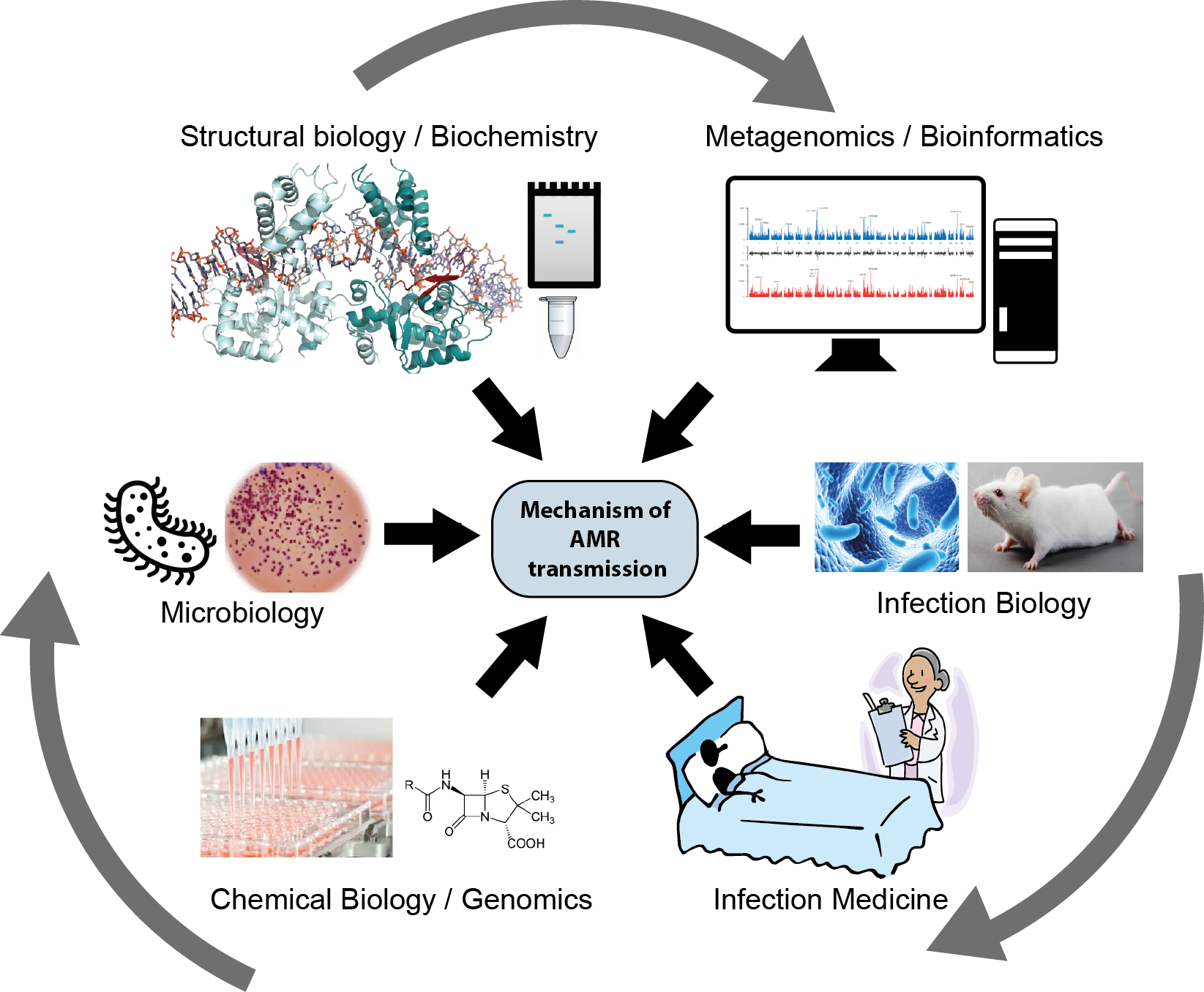
Antimicrobial resistance (AMR) is one of the greatest global health challenges. It spreads rapidly, constantly generating more dangerous bacteria. Mobile genetic elements (MGEs), segments of DNA that can move between bacterial cells, are a major route for resistance transfer in microbial communities. How often such ‘jumping genes’ move, which natural and man-made compounds influence them, and how they move at the molecular level is not understood. In JumpAR, we surveyed MGEs in all bacteria, analysed their resistance cargos and transfer trends, and illuminated the molecular machinery that move them. Drawing on genome and metagenome sequences, we gained a global picture of the abundance and distribution of MGEs and revealed their profound impact on resistance transmission. In a dedicated clinical study, we charted the effects of antibiotics on MGE-driven resistance spreading, and we identified human drugs and environmental compounds that can block AMR gene transfer in bacteria. We further delineated the structure and functioning of the molecular machinery, showing how MGEs build remarkably complex DNA shapes to promote insertion at diverse genomic sites, expanding gene transfer across diverse bacteria. Our collective results vastly expand knowledge on MGE-driven resistance spreading, opening doors to the development of novel strategies against resistance spreading.
- Cell, 2018. Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance
- Current Opinion in Structural Biology, 2019. Jump ahead with a twist: DNA acrobatics drive transposition forward
- Molecular Systems Biology, 2021. Sequence analysis of tyrosine recombinases allows annotation of mobile genetic elements in prokaryotic genomes
- Nucleic Acids Res, 2021. SMART: recent updates, new developments and status in 2020
- The FASEB Journal, 2022. Structure and function of DNA transposition assemblies involved in antibiotic resistance spreading